Filter results
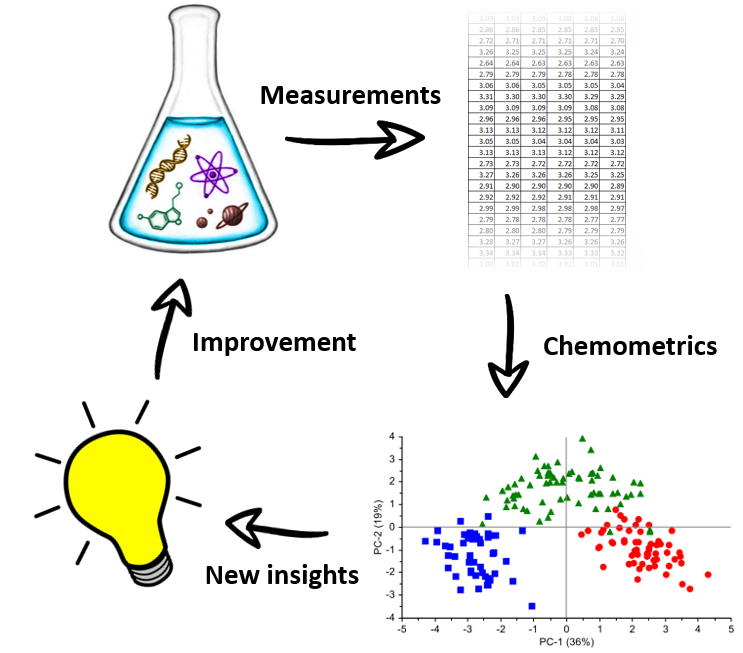
Although continuing technical advances in analytical chemistry allow us to measure complex data with relative ease, analyzing such data is a field of science on its own. Fully exploiting the valuable information you were (or were not) looking for from measured data is exactly the expertise we have at the department for chemometrics. We focus on the development of new or improved data analysis methods, but also on the application of established methods on new types of measurements. Chemometrics finds many practical applications, including but not limited to food safety, healthcare, industrial processing and sustainability. If you would like to contribute to such work, check our website and/or contact us for more information on current projects and internship subjects!
The principal focus of Aquatic Ecology and Environmental Biology is to unravel biogeochemical and ecological key processes explaining the functioning and services of wetland ecosystems - covering freshwater, brackish and marine systems - including the interactions between organisms, surface waters, sediments, groundwaters and the atmosphere. Particular attention is payed to environmental pressures such as enhanced nitrogen deposition, eutrophication and global warming and their effects on the carbon, nitrogen, phosphorus and sulfur cycling. We apply our biogeochemical and ecological insights in the conservation and restoration of wetland ecosystems including lakes and rivers (conservation biology) and in sustainable solutions for water treatment and agriculture. For our research, the analysis of relevant processes in the field situation is combined with experiments in the field, and in the greenhouse, in mesocosms and microcosms both outdoors and in the lab. Several internship possibilities are announced on the website. Tailor made internships related to ongoing projects are often possible. Please contact Bjorn Robroek to discuss whether your interests match with our possibilities.
Peptides display many different characteristics, both structural and functional, based on their amino acid sequence and secondary conformation. In the bio-organic chemistry group we synthesize peptides and try to manipulate their structure and function. These peptides form the building blocks to create new materials for tissue engineering and synthetic vaccines. Moreover, peptides are employed in our group for the design of molecules to diagnose and treat cancer by specifically delivering drugs to diseased cells.
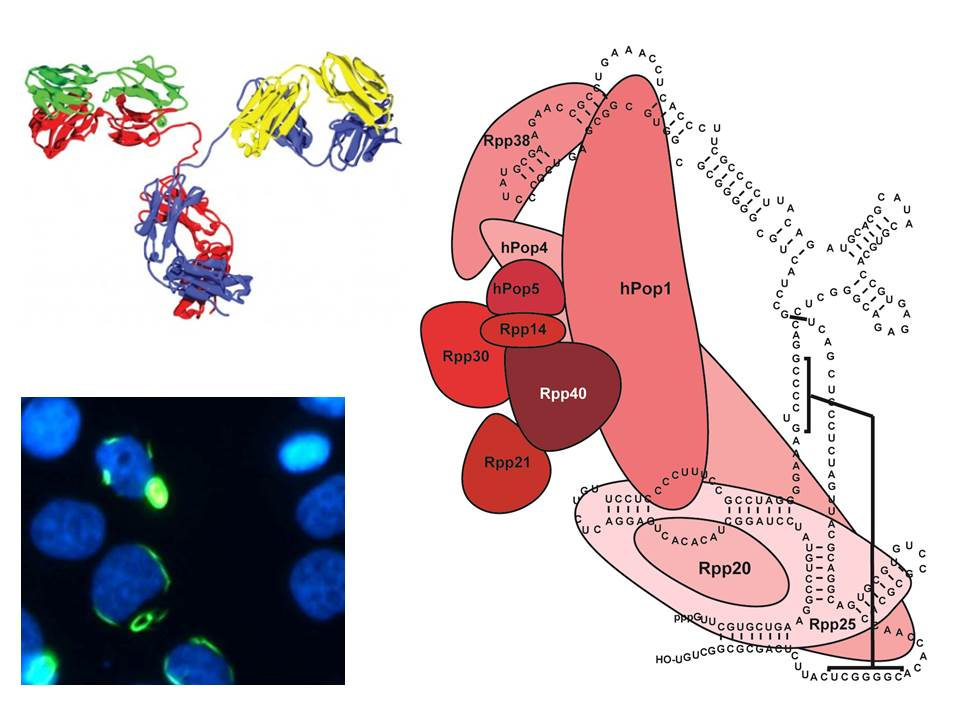
Why do patients with autoimmune diseases mount an 'anti-self immune response'? Genetic, environmental and other factors are known to play a role in the loss of immunological tolerance to self-molecules. However, at the molecular level the underlying mechanisms are only poorly understood. One of the main molecular features of autoimmunity is the production of autoantibodies, often in a very disease-specific fashion and, therefore, they represent biomarkers with diagnostic value. We combine biochemical, cell and molecular biological, and immunological technologies to identify new autoantibody biomarkers, to explore the mechanisms of immune system activation against self-components, with special interest in molecular modifications, and to elucidate structure-function relationships of autoantibody targets, in particular macromolecular complexes involved in gene expression.

What is your blood group? A, B, or O? Did you know that this is determined by complex sugar molecules called glycans? Glycans cover the suface of every cell in the body. They are composed of different carbohydrates that assemble into a dazzling amount of structurally diverse sugar chains. Next to constituting the blood groups, glycans regulate immune recognition and instruct the intestinal microbiome. Moreover, glycan alterations are involved in every major disease.
Our lab uses gene editing (CRISPR) and organoid models to control the glycan structures that cells produce and to recapitulate their functions in health and disease. This enables the production of glycans that have for example beneficial effects on the immune system or the microbiome. Furthermore, this platform allows to engineer “diseased” glycans and to recapitulate their role in pathological processes such as inflammation and autoimmunity and to develop novel glycan-based therapies.
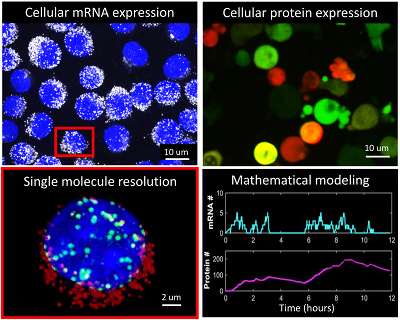
Why do two identical cells look different? Cell-to-cell variability (i.e. noise) in gene expression leads to large differences in mRNA and protein levels in cells. This variability can hamper the treatment of diseases such as HIV and cancer. Due to the architectural complexity of a cell a multitude of factors can be identified that heavily influence reaction dynamics, causing gene expression to deviate from predictable behavior. We combine single-molecule and time lapse imaging with cell-free biochemistry approaches to discern key physical, kinetic, and gene circuit-based factors that determine the outcome of cellular reactions at a single-cell level.
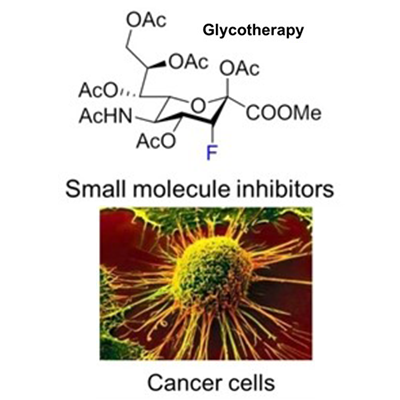
Carbohydrates are the most abundant biomolecules on earth. They play an essential role in biology as a source of energy and regulate many biological processes. We develop new chemistry to synthesize carbohydrate molecules with a variety of applications. The main topics and projects in the group are listed below. Depending on the project we are able to host chemistry, biology or molecular life science internships. Some of the projects are in collaboration with partners from industry.
Stereoselective glycosylation reactions: We develop new chemical methodology to prepare complex carbohydrates via stereoselective glycosylation reactions. To this end, we also study the reaction mechanism of these reactions.
Carbohydrates as drugs: We design and synthesize carbohydrate based drugs toward the treatment of cancer, pathogenic infections and immune deficiencies. Furthermore, we perform cellular tests to evaluate the biological activity of the molecules that we prepare.
Carbohydrate food ingredients and allergies: We synthesize carbohydrate food ingredients and study their health benefits and allergy profiles.
Carbohydrate based personal care products: We design, synthesize and test carbohydrate molecules for the personal care application (cosmetics etc).
If you are interested in one or more of these projects, feel free to contact us for more information
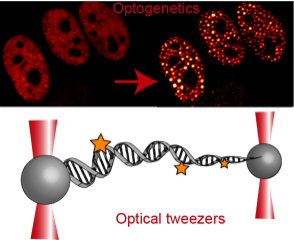
DNA packaging into the nucleus of a cell is a topological organization problem that needs to be precisely regulated, as the genome needs to control the accessibility of the information it encodes. DNA organization occurs on several levels, all involving physical processes. The basic folding of the genome is dictated by principles of polymer physics, active systems driven by molecular motors result in DNA loops, and division into nuclear compartments is regulated by the equilibrium thermodynamics of phase separation. Our lab aims to examine the functional relevance of nuclear architecture and the biophysical mechanisms that drive it. We use a multifaceted and interdisciplinary approach, ranging from cellular- and single-molecule biophysics to engineering.
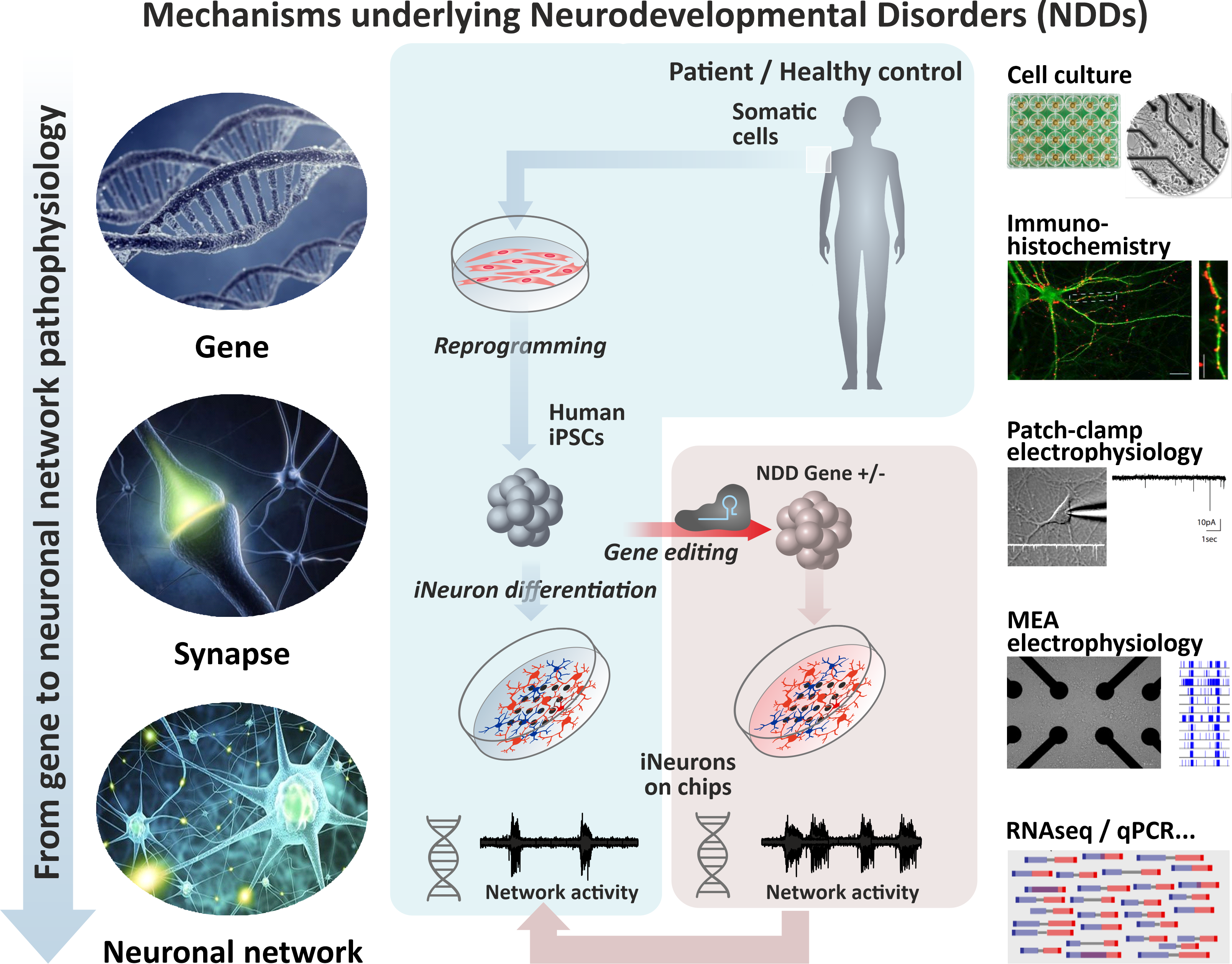
In many neurodevelopmental disorders, including Intellectual Disability (ID) or Schizophrenia, (risk) gene mutations have been identified that directly or indirectly alter neuronal maturation, signaling and neuronal network organization.
In our group for "Cellular Neurophysiology" at the CNS dept, Radboudumc, we are focussing on mutations of epigenetic modifiers that have been found to be causative for ID or Schizophrenia in patients. We aim to reveal the molecular and cellular mechanisms leading to neuronal network dysfunction and to identify new potential targets for therapy.
To this end we make use of neuronal cultures derived from human induced pluripotent stem cells (hIPSCs) obtained from healthy subjects as well as from patients. For our research we combine molecular, neuroanatomical, electrophysiological in vitro techniques (from single cell patch clamp over paired recordings to multielectrode array recordings) in order to reveal the link between gene and neuronal network phenotypes. Internships will be fully integrated in this interdisciplinary research.
The development of chemistry that can be used inside living cells and animals made it possible to visualize and study biomolecules in their native cellular context and greatly contributed to advanced targeted drug delivery strategies. In our research group we are interested in developing such so-called 'bioorthogonal reactions' and use them for specific delivery of therapeutics to cells involved in cancer and autoimmune disease. Furthermore, we are interested in developing molecules that can be used to unveil fundamental biological questions related to autoimmune disease.
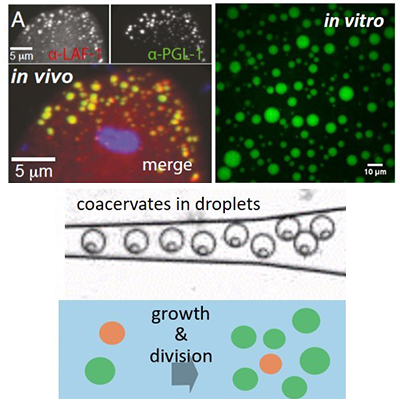
How did the first cell form? In the Soft Interfaces group, we aim to understand how a complex system like a living cell could have emerged from simple organic molecules. By self-assembly of peptides, nucleotides and sugars into liquid coacervate droplets, we make synthetic organelles that grow, fuse, split, and act as microreactors for chemical reactions. Many living cells still bear marks of these coacervate droplets in the form of membraneless organelles. In this case, we use a biophysical chemistry approach to investigate what their function is in cell organization.
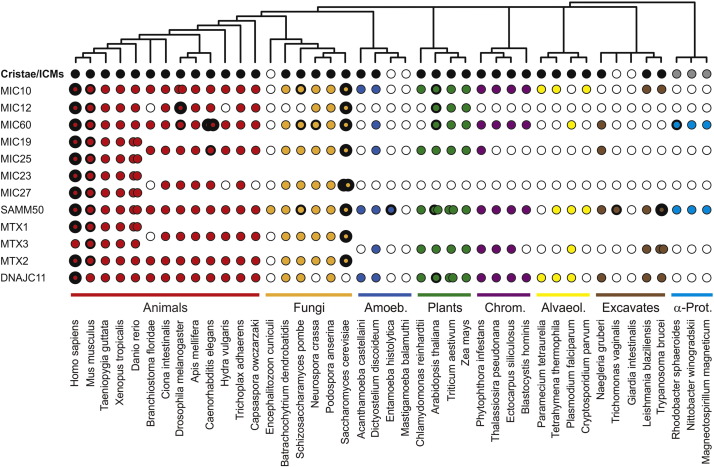
In Martijn Huynen's group at the CMBI we exploit molecular 'omics data to
predict the function of proteins, their interaction in complexes and pathways and their evolution. We focus on biomedically relevant systems like Plasmodium, the mitochondrion and the immune system. We develop methods for 'omics data analysis and collaborate with experimental groups to analyze their data.
We offer internships in the analysis of molecular 'omics data for students with a MLS, Bio, or Medical Biology background. Doing of the course MOL074, Comparative Genomics is highly recommended to an in internship, but can also be done as part of the internship.
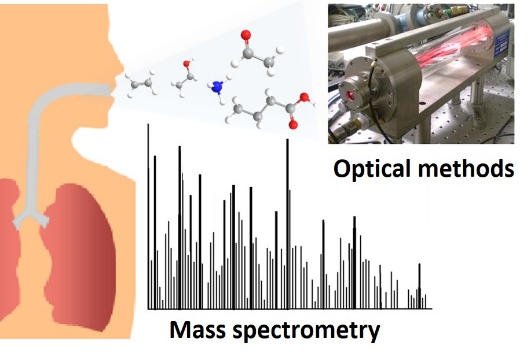
Our group develops and utilizes different detection techniques based on laser
spectroscopy and (real-time) mass spectrometry for monitoring volatile compounds. We perform research in a broad range of applications, from environmental to analysis of biomarkers in human breath. Some of the projects are in collaboration
with colleagues from other departments and/or the academic hospital. We can host physics, science, chemistry, biomedical or molecular life science internships.
Non-invasive detection of biomarkers in human breath: we
develop new methodologies to detect and monitor biochemical processes
in the human body via untargeted and targeted biomarkers in exhaled breath. We combine multivariate analysis tools to relate them to health (diet, exercise, exposure) and diseased status (cancer, infections, etc.).
Sniffing the chemical language of pathogens:We analyze volatile metabolites produced by pathogens as infection-specific biomarkers.
Plasma diagnostics
using spectroscopy: We design, develop and utilize a broadband absorption spectrometer for analyzing discharge plasmas. It is an applied research in the lab, well balanced between physics and chemistry combined with computational modeling and simulation.
Please check our website and fell free to contact us for more information on
current projects and internships.

Development of educational experiments
You are probably familiar with the Education Labs for Molecular Sciences, since most of your lab courses took place here...
The labs facilitate
lab courses for undergraduate students of Chemistry, Molecular Life
Sciences, and Science. An integral part of that job is the development
of experiments that can be used in these lab courses. An educational
experiment for a lab course is different than a research experiment.
Whereas a research experiment is meant to answer a question about
something new and yet unknown, a good educational experiment should:
•
illustrate something that is already known (at least to the teacher
and possibly to the student as well, although results may also surprise
the student at first);
• be known to work and be relatively robust; a student should be able to perform it with a reasonable chance of success;
• be characterized extensively. What are possible catches, what works and what doesn’t?
• teach the students experimental skills.
We offer a limited number of internship positions (no more than two at the same time) in which you can work on the translation of a research experiment into an educational experiment or on the optimization or extension of an existing lab experiment. Having at least some affinity with education (from the standpoint of an educator) is a prerequisite for any internship at our department.
To get more information on the currently available topics please have a look at our webpage!
Improvement of Molecular Sciences courses
We also have good experiences with internships that investigate the teaching of a particular aspect or subject from courses in one of the Molecular Sciences programmes with the objective of improving it. If you are interested in an internship with a 'hands-on' approach to didactics and the teaching of Molecular Sciences, feel free to contact us to discuss possibilities.
Koen van Asseldonk, Luuk van Summeren & Tom Bloemberg
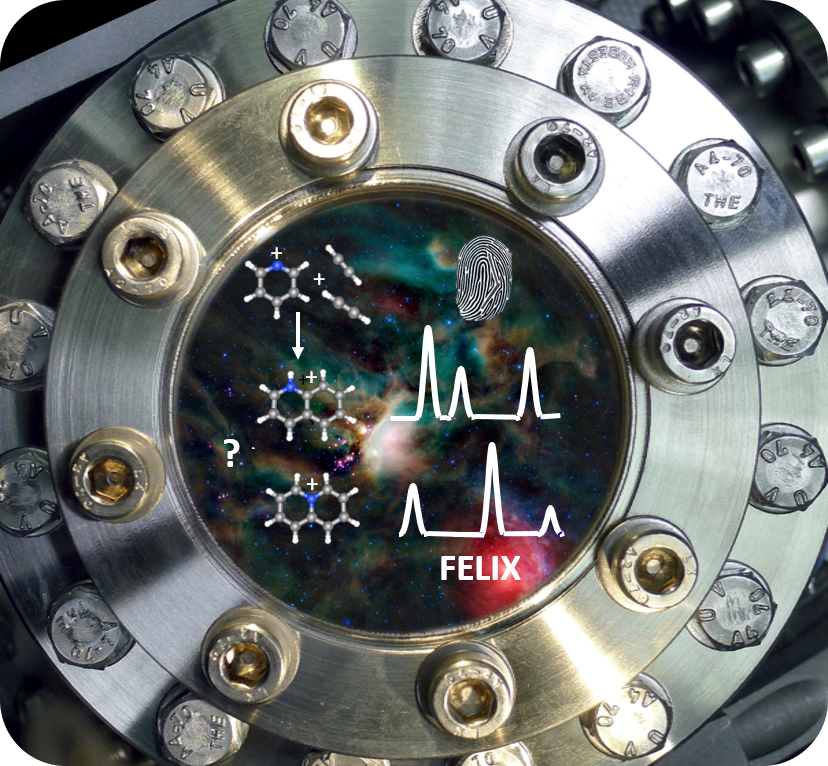 Our group develops and uses mass spectrometric techniques in combination with advanced infrared and terahertz spectroscopy. Our main scientific focus is in the field of astrochemistry. We aim to understand the chemical evolution in astrophysical environments, such as interstellar star-forming regions or (exo-) planetary atmospheres, by simulating their conditions in the laboratory. For this we apply infrared spectroscopy to obtain vibrational fingerprints of molecules to aid their identification in space. The spectroscopic information allows us to identify molecular structures within isomeric mixtures, and thus to unravel and to understand astrochemical reaction kinetics and networks in great detail both in the gas-phase or in ices.
Our group develops and uses mass spectrometric techniques in combination with advanced infrared and terahertz spectroscopy. Our main scientific focus is in the field of astrochemistry. We aim to understand the chemical evolution in astrophysical environments, such as interstellar star-forming regions or (exo-) planetary atmospheres, by simulating their conditions in the laboratory. For this we apply infrared spectroscopy to obtain vibrational fingerprints of molecules to aid their identification in space. The spectroscopic information allows us to identify molecular structures within isomeric mixtures, and thus to unravel and to understand astrochemical reaction kinetics and networks in great detail both in the gas-phase or in ices.
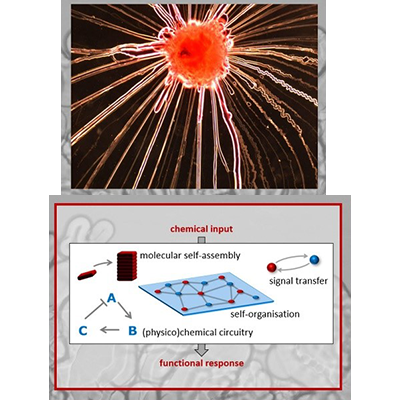
Turning supramolecular chemistry into life-like materials.
We build chemical systems that combine molecular self-assembly and chemical reactions in fascinating life-like behaviour such as growth, shape transformation, chemotaxis and signal transfer.
Currently, we are developing self-assemblies that organize themselves into wire-like structures – in a (primitive) analogy to the growth of slime mold wires. We aim to use these wires to grow a “chemical computer”, where the self-assembled wires “guide” either molecular or electric signals. Ultimately, we envision to use these wires in self-organizing device interfaces that “determine” the path of samples in lab-on-a-chip applications, or neuromorphic electronics that adapt upon growing new connections in the circuit (in analogy to neurons in the brain).
Our research involves a wide diversity of techniques and approaches, such as supramolecular chemistry, systems chemistry, synthesis, electrochemistry, microscopy, spectroscopy, building devices via 3D printing, automated image analysis, etc.
Currently, we have internship projects available on:
- Combining enzymatic reactions and self-assembly for chemical signal transfer along wires.
- Building and programming robotic devices to control the self-organization of our dynamic wire networks.
- Using electrochemistry for controlled assembly of dynamic, conductive connections in neuromorphic circuits.
- Design and synthesis of molecular building blocks for new concepts in out-of-equilibrium self-assembly to control the growth of the networks.
Depending on your preferences, we will design a project that suits your interest and background. For more info on our research and contact details, please check out our new website: www.korevaarlab.com.
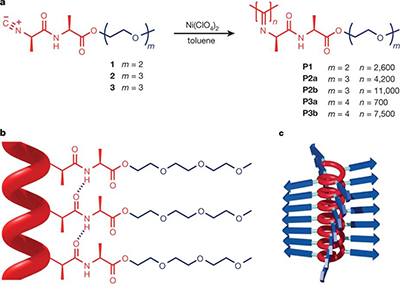
Life is supported by fibrous hydrogels: they are present inside our cells (cytoskeleton) and between them (extracellular matrix). Synthetic gels, typically have very different properties than gels of biological materials, such as actin, fibrin and collagen, and, therefore are often unsuited for biomedical applications.
In the Molecular Materials group, we develop synthetic materials that do behave like biological hydrogels. The research in our group follows two lines: gel design, e.g. a gel that becomes 10-fold stiffer by heating 1 °C and biological application of the gel, for instance as an advanced cell culture medium or even for medical applications.
For internships, we host students from Chemistry and Science (most often in gel design, analysis and modelling) as well as students from Molecular Life Sciences and Medical Biology (for biomedical studies and cell biology).
CURRENTLY, WE ARE LOOKING FOR STUDENTS FOR CELL CULTURE APPLICATIONS. Interested, shoot us an email: p.kouwer@science.ru.nl
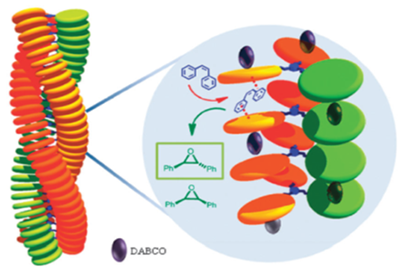
Molecular machines and motors in metal-based catalysis
Inspired by the complex working mechanisms of natural enzymes and biological motors and machines, our group develops supramolecular systems that can act as catalysts to convert (polymeric) substrates into desired products
with a high level of control.
In particular, we aim at developing a new technology to write, store, and
read information onto molecules, i.e. on single polymer chains, with the help of molecular machines that are inspired by the Turing
machine, ahypothetical proposed by the British mathematician Alan Turing in 1936 as
the general basis for the operation of a computer.
Data storage on polymer chains is highly relevant nowadays as it may
help solve the problem of handling the exponential growth of information
that is currently trafficking the internet.
Moreover, molecular data storage is expected to cost orders of magnitude less energy than conventional data storage in large data centers.
The group is working on the encoding of digital information into single polymer molecules in the form of chemical groups, such as (R,R)- and (S,S)-epoxides or (R)- and (S)-sulfoxides, which represent the digits 0 and 1. These are imprinted with the help of light-switchable catalytic machines that threads onto a polymer chain containing alkene double bonds or aryl sulfides. While moving along the chain the catalytic machines (ep)oxidize these functional groups (Figure). The enantioselectivity of the oxidation reactions is controlled by metal-containing catalysts of which the chirality can be switched by an attached Feringa-type molecular motor.
The work involves the synthesis of chiral cage compounds, light-switchable metal-containing catalysts and polymers, and the study of their properties with various techniques (NMR, IR, circular dichroism, UV-vis, fluorescence), threading experiments, and catalysis experiments, all focused on the writing of information.
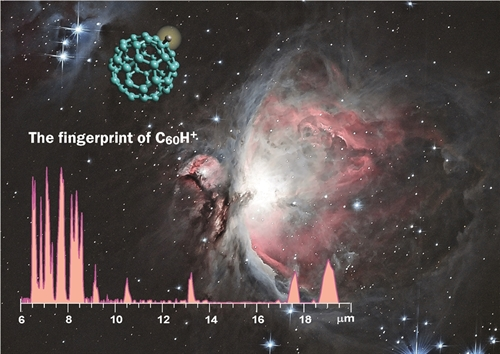 Our group combines and integrates mass spectrometry with IR spectroscopy, enabling us to obtain infrared spectral fingerprints for mass-selected ions inside the mass spectrometer. We apply infrared ion spectroscopy in various analytical challenges of identifying molecular structures of low-abundance compounds within complex mixtures, e.g. in biomarker discovery for inborn errors of metabolism. In more fundamental studies, we investigate molecular spectra and structures of ionized molecules, e.g. to pin down the molecular structure of product ions in tandem mass spectrometry or to obtain laboratory IR spectra for molecular ions that are suspected to occur in astrophysical environments.
Our group combines and integrates mass spectrometry with IR spectroscopy, enabling us to obtain infrared spectral fingerprints for mass-selected ions inside the mass spectrometer. We apply infrared ion spectroscopy in various analytical challenges of identifying molecular structures of low-abundance compounds within complex mixtures, e.g. in biomarker discovery for inborn errors of metabolism. In more fundamental studies, we investigate molecular spectra and structures of ionized molecules, e.g. to pin down the molecular structure of product ions in tandem mass spectrometry or to obtain laboratory IR spectra for molecular ions that are suspected to occur in astrophysical environments.
 Nanomedicinal approaches can be utilized to overcome existing challenges in medicine that cannot be addressed by traditional pharmaceutical approaches. Macromolecules are an essential component of nanomedicine, not only for the construction of nano-devices but also for the decoration of such devices with chemical functionalities.
Nanomedicinal approaches can be utilized to overcome existing challenges in medicine that cannot be addressed by traditional pharmaceutical approaches. Macromolecules are an essential component of nanomedicine, not only for the construction of nano-devices but also for the decoration of such devices with chemical functionalities.
Antibiotic resistance is a major problem in medicine and poses a threat to global human health. To better understand the molecular processes underlying resistance and ultimately find a solution to this problem we synthesize and apply small molecule tools. We use multistep organic synthesis to develop fluorogenic substrates for key enzymes involved in the emergence of resistance and apply them to live cells to unravel the molecular mechanisms that lead up to resistance. Furthermore, we have a special interest in the roles of nucleic acids (RNA and DNA) in the development of antibiotic resistance. We synthesize potential antibiotic molecules that target these nucleic acids in bacteria. Ultimately, we use our findings to device new molecular strategies to combat the antibiotic crisis.
The ultimate aim of our research is to understand how life works and where it comes from. Research in our group is multidisciplinary and exploits microfluidic tools to create synthetic cells or to quantify gene expression product in single cells. We have a strong interest in complex molecular systems and study how networks of chemical reactions create functional behavior like oscillations, switches or amplifiers. In effect, we try and construct molecular computers. We now try to identify how minimal complex systems can arise from mixtures of coupled reactions, and, ultimately, how life arose out of chemical reaction networks of increasing complexity.
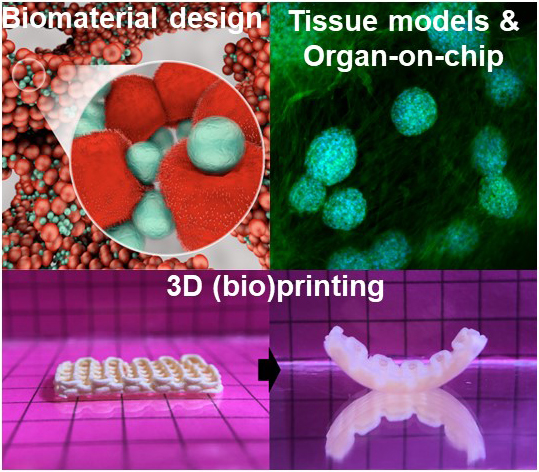
Are you excited about research at the interface of materials, chemistry, biology and medicine? We are a multidisciplinary and international group with diverse scientific and clinical backgrounds. We strive to:
Interested to learn more about current projects? Shoot us an email at mani.diba@radboudumc.nl
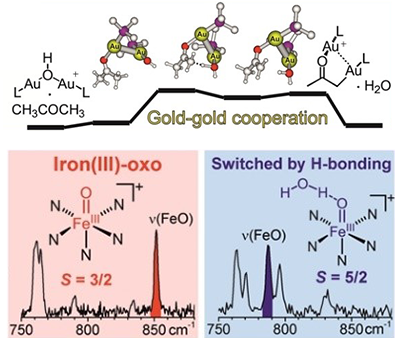
The Roithová group is studying and designing metal complexes to mediate efficient chemical transformations. We seek inspiration from enzymes that can make reactions selective and can use abundant reactants such as oxygen or CO2. By researching working principles of enzyme-inspired catalysts and by coupling these catalysts with photochemistry and electrochemistry we aim to develop more efficient chemical transformations and thereby to contribute to reducing the environmental impact of chemistry in the future.

Our structural bioinformatics group at the CMBI is led by Li Xue and Hanka
Venselaar. Our research interests focus on bridging artificial intelligence (AI) and structural biology to better understand the molecular basis of diseases and to rationalize drug design. We use information obtained from available structures,
homology models, and other data-sources in order to, for example, interpret variations, improve experimental design, drug docking, etc.
One of our main projects focusses on developing AI methods for better cancer vaccine design. A combination of Deep Learning and Integrative Modelling is used to predict MHC-peptide-T cell receptor complexes. We welcome talented students to join our endeavor.
We offer internships for Chem/MLS/Biol students (both Ma and Ba) either with a clear interest and understanding of protein folding or with an enthusiasm of AI.
An internship without programming experience is possible in the field of 3D modelling and mutant analysis. The AI project requires at least a decent basic level of programming skills. Internships, also in collaboration with other departments, are possible in many different fields.

The Rutjes group focuses on the development of new synthetic methodology, mainly focused on the synthesis of small organic molecules with specific biological activity. This includes projects on chemical reactions in continuous (photochemical) microreactors, medicinal chemistry projects to prepare lead compounds for specific drug targets, and asymmetric synthesis methods to prepare enantiopure products.
In nature, countless complex structures are known, and mimicking these structures can be a challenging task. How do you design a nano- or micro-system from the bottom up? Our goal is to design functional supramolecular structures and apply them to advance the field of nanomedicine.
Our group finds their inspiration in natural materials and processes. It is our aim to develop functional polymers, peptide and protein-based hybrid materials with biological activity. By using a variety of synthetic techniques, such as controlled polymerization, peptide synthesis and protein engineering methods. We furthermore mimic natural biological processes by compartmentalization and assembly of biocatalysts in polymeric capsules (polymersomes) for the design of synthetic mobile systems.
 In Peter-Bram ’t Hoen’s group at the Center for Molecular and Biomolecular Informatics, we develop computational approaches to advanced personalized medicine. We integrate molecular -omics (genome sequencing, RNA-seq, proteomics, metabolomics) and clinical data to study rare disease mechanisms, identify drug targets, understand heterogeneity in disease course and onset and stratify patients for therapies.
In Peter-Bram ’t Hoen’s group at the Center for Molecular and Biomolecular Informatics, we develop computational approaches to advanced personalized medicine. We integrate molecular -omics (genome sequencing, RNA-seq, proteomics, metabolomics) and clinical data to study rare disease mechanisms, identify drug targets, understand heterogeneity in disease course and onset and stratify patients for therapies.